So this is stolen from the brilliant Aaron Tay, but I have adapted it here for NHS librarians specifically and step 5 is a bit different from in his post as I think the Unpaywall API changed since he wrote it. Thank you so much Aaron for your brilliant work! And also thanks to Liam Bullingham who pointed me towards the very helpful and informative blog post.
1. Get a list of your organisation’s publications for a given year in a format that includes DOI. For the purposes of this example we will use Pubmed but do be aware that this does miss out some nursing and allied health publications which may be on CINAHL instead. (1)
On Pubmed Advanced Search, use the AFFILIATION and DATE – PUBLICATION fields to identify papers published by authors from your organisation in a particular year. This is what my search looked like, you will need to change the info in the ‘Affiliation’ bracket to match your organisation.
((“2020/01/01″[Date – Publication] : “2020/12/31″[Date – Publication])) AND (“royal marsden”[Affiliation])
I had 996 results. I clicked on Save and downloaded them all in CSV format.
At this stage, you can use Excel to delete some of the lines of the CSV and create a ‘test’ CSV file with only 5-10 lines of data instead of hundreds! This is because the checking process can take a few hours if it’s processing hundreds of lines and it will be much quicker to check a few first to see if your code is correct, before asking it to do the whole lot.
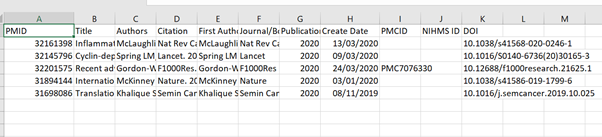
2. Open your CSV file in OpenRefine
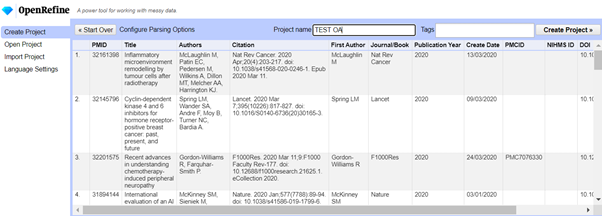
Run the OpenRefine software. I use the 3.4.1 version with Java and it opens in Google Chrome with some weird background program running at the same time. The first screen you see should give you the option to Get data from > This computer > Choose Files. Open the Test CSV file that you have created. Give your project a name, leave all the other options unchanged, and then click on Create Project.
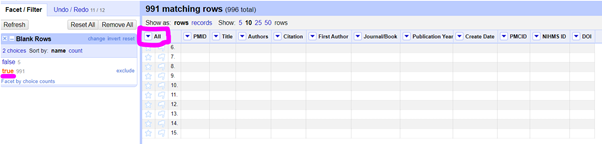
3. Delete blank rows if they are there (Optional)
At this stage although I thought I had deleted several hundred rows in Excel to create my TEST spreadsheet with only 5 lines, they were all still there, just… blank. You can delete these blank rows by doing this:
Click on the little arrow next to the ALL column and select ‘Facet by blank (null or empty string). This allows to you select only the blank rows if you click on the relevant facet on the left. Then you can click on the little arrow next to ALL again, select EDIT ROWS > REMOVE MATCHING ROWS to delete the blank ones. When you’re done, click on the little X next to Blank Rows on the left to close the Facet.
4. Run the Unpaywall DOI
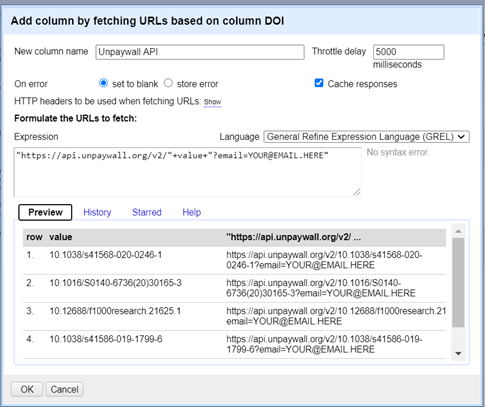
Click on the little arrow next to the DOI column that Pubmed has created for us. Click on Edit Column > Add column by fetching URLs
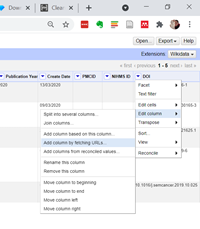
A popup box will appear. Copy and paste this code into the Expression box, and give the new column the name UNPAYWALL API. Edit the info after the email to be your real email address! It doesn’t matter what address you use, it won’t send you any spam or anything, it’s just for validation.
“https://api.unpaywall.org/v2/”+value+”?email=YOUR@EMAIL.HERE“
When you’re ready, click OK and wait! It can take several hours if you’re doing a few hundred records, so go away and do something else if this is the case. If you’re only doing a test document with 5-10 it should take only a few minutes.
What is the OpenRefine software doing? It is talking to the Unpaywall API, checking the info for each doi, and reporting back what it finds there.
5. Pull out the relevant information from the Unpaywall data
So you should now see a new column appear that has lots of complicated info! This is everything that the Unpaywall API has reported back in a format called JSON, and lots of it is not relevant to our question of whether or not the paper is Open Access or not. The only bit that is important is the bit that says “is_oa” – this is set to either TRUE or FALSE.
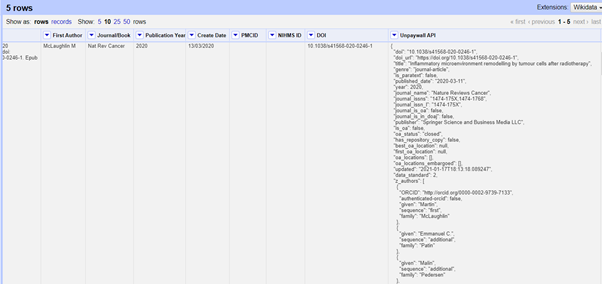
To get only the “is_oa” information out, we need to create yet another column. Click on the small arrow at the top of your new UNPAYWALL API column, then select EDIT COLUMN > ADD COLUMN BASED ON THIS COLUMN. In the popup box that appears, give your new column the name IS OA and type this into the EXPRESSION box: value.parseJson().is_oa
Then click OK.
6. Analyse your data
So all the papers that say TRUE in the IS OA column are Open Access, and all the ones that say FALSE are not freely available to read! Click on the little arrow next to the IS OA column and select FACET > TEXT FACET to quickly identify how many are true and how many are false. Then it’s simple maths to work out what the % of Open Access papers are – for my institution, of the 996 papers published in 2020 that are indexed on Pubmed with a DOI, 671 were available Open Access (67.3%). There were 3 that Unpaywall didn’t have information on – these showed as ‘blank’ in the IS OA column. You can also do further analysis on what type of Open Access they are if you repeat step 5 above with the code value.parseJson().oa_status
If you used a test spreadsheet you can try and repeat steps 2-5 above with the full dataset – just be prepared to wait a few hours for the results when doing step 4.
Any problems or questions do let me know! I will do my best to help – yiwen.hon@rmh.nhs.uk
(1)If you have used sources other than PubMed for your list of publications then you might need to do some extra cleaning to get the DOI info onto its own column. For example, I do searches on CINAHL as well as Medline/Pubmed and deduplicate on Endnote. I usually export as a big bibliography in APA format and end up with lots of references in this format:
Aggelis, V., & Johnston, S. R. D. (2019). Advances in Endocrine-Based Therapies for Estrogen Receptor-Positive Metastatic Breast Cancer. Drugs. 79, 1849–1866. doi:10.1007/s40265-019-01208-8
From this, you can get a column just with the DOI information by doing this: Edit column > Add column based on this column > value.split(“doi:”)[-1] (I got this from https://stackoverflow.com/questions/47088117/extract-text-after-a-string-using-grel)
Another very useful bit of code for getting rid of punctuation at the end of cells is: replace(value, /[.]$/, ”) I don’t understand this code much but it works! So if you have a doi that is wrongly written as 10.1007/s40265-019-01208-8. it will become 10.1007/s40265-019-01208-8
Only the full stop at the end of the data is removed, not the full stop at the beginning. You can also replace the bit in between the square brackets [.] with other punctuation, like commas. I got this from https://groups.google.com/g/openrefine/c/9LB9OT0q2rY
Pingback: OpenRefine for NHS Librarians | The Adventures of YiWen the Librarian
This is fantastic! Thanks so much for these posts! I think the idea of coding is very scary to me, but to find realistic useful examples of what it can do is brilliant and breaks down the barrier a little bit!